Overview
Introduction
A recent conversation on Github led to an ontological problem. You might consider that a pretty heavy outcome for a pretty pedestrian discussion about typos, but here we are nonetheless. So here’s the question: What, fundamentally, is the nature of bootstrapping? While most statisticians have long memorized the standard bootstrapping algorithm and some of its applications, deviations from the standard paradigm are explored less often. What happens if we don’t sample all of the data? What happens if we don’t use replacement?
The short answer is that both can lead to useful results, given some important caveats and exceptions. We’ll explore them today.
The theory of bootstrapping
The standard algorithm
As always, let’s start with some theory. The standard bootstrapping algorithm roughly follows:
- Determine a statistic of interest derived from a particular sample of length n
- Using the previous data, generate new data (also of length n) by sampling with replacement
- Calculate the sample statistic using this data
- Repeat k times (often 1,000 or more) to generate a distribution of the sample statistic
- With this distribution, calculate bias, confidence intervals for the statistic, etc.
The confidence intervals are usually the most interesting estimate that we can derive from the method. Getting the sampling statistic itself isn’t that big of a deal, and we don’t need a complicated sampling algorithm to reproduce what we already have. On the other hand, it can often be difficult to get confidence intervals. Estimating confidence intervals for sample medians, for example, can get really hard.
Luckily, we have many, many options for calculating confidence intervals from the bootstrapped distribution. The boot package in R provides five. A couple of approaches stick out:
- Use the variance of the bootstrap distribution for a normal or t distribution approximation.
- Use percentiles. A 95 percent confidence interval is based on the 2.5 and 97.5 percentiles.
This works in most cases, with some notable assumptions:
- Your original sample data is representative of the population data
- The bootstrap distribution is roughly symmetric
- The distribution of your data has finite moments
- Your statistic is not at the extreme of the distribution of the data
Even when these assumptions are violated, there is usually some sort of correction available. Different statistics, like trimmed means, can be used for distributions that don’t have finite moments. Special sampling algorithms handle extreme-valued statistics. The list goes on and on.
Why does this work?
Bootstrap estimation rests on a couple foundational principals in mathematical statistics. Consider perhaps the most basic problem in estimation, sample means. Since sampling statistics are based on random samples, they are also random variables, with their own distributions. My estimate of the average height of men in America is different for every sample of men I use.
Just because the sampling statistic is random doesn’t mean that there isn’t a whole lot we can say about it. In fact, thanks to the Central Limit Theorom (CLT), we know know that the sample mean $\bar{x}$ is an unbiased estimator of the population mean $\mu$ that it follows a particular normal distribution. More precisely,
In English, we say that the sample mean converges in distribution to that described above. Convergence in distribution can seem a little arcane, but it’s actually pretty straightforwad. Given a random variable whose distribution function depends on n, convergence in distribution implies that
For example, the classic normal approximation to a binomial random variable is based on the fact that a binomial random variable converges to a normal random variable as $n \to \infty$. The more we increase n, the smoother the distribution of the binomial random variable becomes.
If you are familiar with Moment Generating Functions, you can follow a proof of the CLT. I’ll skip it today, since many other people have done a much better job than I can.
Just as the sample mean converges in distribution to a particular distribution, we can use the statistic’s empirical distribution function (EDF) to estimate the true distribution. Moreover, given enough observations, we know that EDF’s converge almost surely to true distributions.
So that’s what we’re doing when we bootstrap. We take a particular sampling statistic and generate its empirical distribution. With that, we can make statements about the true distribution of the statistic. As we make more random draws, we get a better approximation. And we can keep going as long as we’re willing to let our computer run.
Thanks to the CLT, we know exactly what kind of distribution we are approximating when working with sample means. The variance of the bootstrap samples is the standard error of our estimate. This is the key piece that allows us to calculate the confidence intervals described above.
Deviations from the standard procedure
As Geyer points out, when bootstrapping, we are sampling from an approximation of the sample statistic’s distribution. As we discussed above, even though this is incorrect, we approach the true distribution with enough iterations. But in an ideal workld, we would prefer sample from the true distribution. Subsampling can do this:
- Determine a statistic of interest derived from a particular sample of length n
- Using the previous data, generate new data (of length $b « n$ ) by sampling without replacement
- Calculate the sample statistic using this data
- Repeat k times (often 1,000 or more) to generate a distribution of the sample statistic
- With this distribution, calculate bias, confidence intervals for the statistic, etc.
By sampling without replacement, subsampling utilizes the correct distribution, but the sample statistic that we generate is based on the wrong size. This has several ramifications. First and foremost, our sampling statistic is biased.
Fortunately, since $n$ and $b$ are known, we can correct for this.
And
This is approximately true for any sampling statistic. More importantly, as Poltis, Romano and Wolf this method converges at a first-order rate. While the bootstrap converges faster (second-order convergence), subsampling can work in a variety of situations where bootstrapping fails. A notable case is time series, where autocorrelation screws up our bootstrapping algorithm.
Granted, we can also sample less than the full length of the vector while keeping replacement. This methods is known as the m out of n bootstrap, and it is often credited to Bickel, Gotze and van Zwet. They note that there are some cases where this method can work where the standard algorithm doesn’t, but we pay for this flexibility in efficiency.
Experimenting with subsampling
Calculating confidence intervals
Let’s see how these alternative algorithms work in practice. In particular, given a case where standard bootstrapping works, how do these alternatives compare against each other? The easiest case is to work with means and their confidence intervals. Our experiment will compare biases and convergence rates for subsampling and m out of n bootstrapping by measuring the algorithm’s confidence interval with a “true” interval parametrically derived. This true interval is easy to find when working with means.
We would like our experiment to address a few different questions:
- Which methods converge most quickly?
- What about the subsampling rate?
- Do data-specific parameters, like its size, mean and variance affect convergence?
To do this, I’ve added a new set of functions to the blog’s sister package. Let’s walk through them.
First things first, we need some methods for generating confidence intervals. The most basic approach follows the definition of corrected standard errors described above. We then get the confidence interval with a normal approximation.
red_se <- function(x, probs = c(.025, .975), n, b, xbar) {
se <- sd(x) * sqrt(b / n)
qnorm(probs, xbar, se)
}For a quantile-based approach, we borrow some code from Geyer.
ss_quantiles <- function(x, probs = c(.025, .975), n, b, xbar) {
z.star <- sqrt(b) * (x - xbar)
qntls <- quantile(z.star, probs[2:1])
xbar - qntls / sqrt(n)
}The first line, calculating z.star gives us an approximate distribution based on the subsamples. Here, x is the vector of subsampled statistics, using shorter data, while xbar is the sample statistic based on a sample of length n. The next step draws quantiles from this adjusted distribution, and the final line returns the corrected confidence interval.
While we don’t typically go through all of the work of generating the EDF, it’s still possible in a relatively small number of steps. The formula for the corrected EDF, based on a smaller sample size, comes from a post on Larry Wasserman’s amazing (and sorely missed) blog.
First, a function for the EDF.
emp_dist <- function(t, q, ss_stat, samp_stat, b) {
id <- sqrt(b) * (ss_stat - samp_stat) <= t
mean(id) - q
}The mean of id is the EDF for a vector of subsampling stats. t is the probability threshold from all distribution functions, i.e. $F(t) = P(X <= t)$. I added the parameter q, because we need to find the root of this function for a particular quantile value.
qntl_solve <- function(q, ss_stat, samp_stat, b, n, int) {
root <- uniroot(emp_dist, int, q, ss_stat, samp_stat, b)$root
samp_stat + root / sqrt(n)
}The uniroot function searches an interval int for the root of a function. It solves $f(x) = 0$. The subtraction in the previous function allows us to solve $f(x) = q$ or $f(x) - q = 0$. The expression samp_stat + root / sqrt(n) generates one bound of the confidence interval. We need a vectorized version to get both at once.
quantile_ev <- function(x, probs = c(.025, .975), n, b, xbar, int = c(-500, 500)) {
vapply(probs, qntl_solve, numeric(1), x, xbar, b, n, int)
}These functions handle the confidence intervals for subsampled statistics. Similar functions were created for bootstrap confidence intervals. They will be our benchmark.
Functions for simulation
A generic bootstrap simulator follows.
bs_sim <- function(x, bsi, replace, funs, n, b, ..., probs = c(0.025, 0.975)) {
# Generate random sample estimates
ids <- rerun(bsi, sample.int(n, size = b, replace = replace))
estimates <- map_dbl(ids, ~ mean(x[.x]))
# Calculate confidence intervals
all_args <- list(list(x = estimates, probs = probs, n = n, b = b, ...))
ints <- invoke_map(funs, all_args)
# Generate results
res <- invoke(rbind.data.frame, ints)
res_nmd <- set_names(res, c('lwr', 'upr'))
nmd <- add_rownames(res_nmd, var = 'method')
nmd$replace <- replace
nmd
}Here are the arguments to the function:
xis a vector that we are resamplingbsiis the number of bootstrap iterationsreplacecontrols whether we use replacementfunsis a list of different functions for calculating confidence intervals
The remaining parameters concern sampling size and quantiles for intervals. The function first produces a series of ids for indexing our data vector x. With these indexes, we subset our data vector and calculate the mean, our sample statistic to bootstrap. The next two expressions calculate the confidence intervals using all the functions we’ve provided. Finally, we produce a data frame of the resulting confidence interval from each method.
For convenience’s sake, we wrap this function in compare_methods, which is there to correctly pass all of the parameters to our bootstrap tool. It also compares the resulting confidence intervals against a “true” interval derived from the normal distribution.
compare_methods <- function(param, funs) {
# Params
n <- param$n
b <- n * param$r
nms <- c("lwr", "upr")
# Simulated vector
x <- rnorm(n, param$mu, param$sigma)
# Get the sample standard error and quantile
xbar <- mean(x)
se <- sd(x) / sqrt(n)
true <- map(c(lwr = .025, upr = .975), qnorm, xbar, se)
# Simulation
sim <- bs_sim(x, param$bsi, replace = param$replace, funs, n = n, b = b, xbar = xbar)
# Output
param_cbm <- c(param, true = true)
param_rep <- do.call(rbind.data.frame, rerun(length(sim[[1]]), param_cbm))
cbind(sim, param_rep)
}The output of this function is a combination of each method’s confidence interval, as well as the parameters used to generate the simulation.
Last but not least, we need a tool for generating our experiment. This will produce all of the different combinations of the parameters that we wish to try, and it will handle iteration. Here it is.
bootstrap_experiment <- function(funs,
nrange = c(1000, 10000),
rrange = c(.25, .75),
mus = c(0, 4.5),
sigmas = c(1, 5.5),
bsi_range = c(1000, 10000),
length.out = 5,
niter = 5) {
# Either a single shared length for each range of params, or individual lengths
if (length(length.out == 1)) length.out <- rep.int(length.out, 5)
# Create ranges of values
params <- list(n = nrange, r = rrange, mu = mus, sigma = sigmas, bsi = bsi_range)
ranges <- map2(params, length.out, ~ seq(.x[1], .x[2], length.out = .y))
ranges$i <- seq_len(niter)
ranges$replace <- c(TRUE, FALSE)
# Create all test combinations
combinations <- cross_n(ranges)
# Execute all tests in parallel
tests <- mclapply(combinations, compare_methods, funs, mc.cores = 4)
out <- bind_rows(tests, .id = "test")
mutate(out, bias.lwr = (lwr - true.lwr), bias.upr = upr - true.upr,
bias.sqr = sqrt(bias.lwr^2 + bias.upr^2))
}We build the experiment from a set of ranges:
- The number of observations in our original sample
- The proportion of the data used in the subsample
- The true mean of the first sample
- Its true variance
- The number of iterations to be used in bootstrapping
The argument length.out control the number of value we pull from the ranges. By default, we take five values over even intervals. For each combination of parameters, we have five replicants. This is controlled by the final argument, niter.
All of the combinations of test parameters are stored in a big list. We 93,750 different combinations, using the default parameters. This will require thousands of samples for each combination. For that reason, we iterate through this list in parallel, using mclapply. To measure performance, we check for bias at the upper and lower ends of the confidence interval. I combined these separate biases with the euclidean norm.
Putting our code to work
A quick sanity check
The preceding functions are applied in a script available here. Please don’t source the script or run the whole thing at once. Running the bootstrap experiment takes a long time. We’re talking a few hours of computation on my newish MacBook Pro. For that reason, I’ve also included one example test run. That’s here.
Let’s start just by comparing confidence intervals from a variety of sampling schemes. I know. I know. We’ve put the cart in front of the horse. Nonetheless, it’s useful to see the simpler functions in action before running through all of the interations of our experiment. We’ll generate confidence intervals in the following:
- Estimates using the standard bootstrap
- Estimates using a subsample, with replacement. These are biased when we don’t use the corrected functions.
- Estimates using a subsample, with replacement and the corrected functions.
- Estimates using a subsample, without replacement and the corrected functions.
The third option above is m out of n bootstrapping, while the fourth is standard subsampling. Here is how we call all four methods.
# Comparing BS sampling schemes -------------------------------------------
n <- 1000
b <- n * .25
x <- rnorm(n, 4, 2)
mu <- mean(x)
# Actual values
se <- sd(x) / sqrt(n)
true <- c(lwr = .025, upr = .975) %>% map(qnorm, mean(x), se)
# CI functions
full_funs <- list(se = se_ci, qntls = qntl)
red_funs <- list(se = red_se, ss_qntls = ss_quantiles, emp_qntls = quantile_ev)
# Bootstrapped values
full <- bs_sim(x, 10000, replace = TRUE, full_funs, n, b = n, mu)
biased <- bs_sim(x, 10000, replace = TRUE, full_funs, n, b, mu)
red_rep <- bs_sim(x, 10000, replace = TRUE, red_funs, n, b, mu)
red_nrep <- bs_sim(x, 10000, replace = FALSE, red_funs, n, b, mu)The following table compares the confidence interval from each bootstrapping method with the parametrically-defined confidence interval for a mean.
# Test values
tests <- list(full, biased, red_rep, red_nrep) %>%
set_names(nm = c("full", "biased", "adjusted, rep", "adjusted, no rep")) %>%
bind_rows(.id = "type") %>%
mutate(bias.lwr = (lwr - true$lwr),
bias.upr = upr - true$upr,
bias.sqr = sqrt(bias.lwr^2 + bias.upr^2))
knitr::kable(tests[-5], digits = 4)| type | method | lwr | upr | bias.lwr | bias.upr | bias.sqr |
|---|---|---|---|---|---|---|
| full | se | 3.8418 | 4.0944 | -0.0022 | 0.0025 | 0.0033 |
| full | qntls | 3.8427 | 4.0940 | -0.0012 | 0.0020 | 0.0024 |
| biased | se | 3.7175 | 4.2192 | -0.1265 | 0.1272 | 0.1794 |
| biased | qntls | 3.7205 | 4.2201 | -0.1235 | 0.1282 | 0.1780 |
| adjusted, rep | se | 3.8443 | 4.0917 | 0.0003 | -0.0003 | 0.0004 |
| adjusted, rep | ss_qntls | 3.8444 | 4.0943 | 0.0004 | 0.0023 | 0.0023 |
| adjusted, rep | emp_qntls | 3.8416 | 4.0916 | -0.0024 | -0.0004 | 0.0024 |
| adjusted, no rep | se | 3.8606 | 4.0754 | 0.0166 | -0.0166 | 0.0234 |
| adjusted, no rep | ss_qntls | 3.8593 | 4.0763 | 0.0153 | -0.0157 | 0.0219 |
| adjusted, no rep | emp_qntls | 3.8596 | 4.0767 | 0.0156 | -0.0153 | 0.0219 |
Just as we expected, using a subsample introduces quite a bit of bias in our confidence interval. Nonetheless, the corrected functions essentially remove this bias. When we don’t sample with replacement, convergence for the subsampling method seems to be an order of magnitude slower. Again, this is what we expected.
A short but not meaningless detour into convergence
The previous section begs a question: What do we mean when we claim that an algorithm is “first-order” or “second-order” convergent? Let’s be honest, numerical analysis and the analysis of algorithms aren’t exactly popular classes among Statistics deparments.1
To paraphrase something from Wikipedia, an algorithm “converges” when its error is below a pre-defined threshold, and the rate of convergence is the speed at which it approaches this threshold. Measuring “approaching” can be a bit tricky, but in many statistical algorithms the rate depends on the number of observations or iterations used.
In resampling algorithms, we are interested in iterations. In other words, as we increase the number of iterations in the algorithm, our error should reduce at a particular rate. The formula for calculating the rate is relatively simple. To start, assume we have a process that generates a sequence of estimates $x_k$, where $k$ is the current iterations. Let $\varepsilon_k = \left|x_k - \mathrm{L} \right|$ be the error term as we approach the true value $\mathrm{L}$. Then, we define convergence as
where $\mu$ is some constant value greater than 0. Our convergence rate depends on $q$. If $q = 1$, our convergence rate is linear or first order. If $q = 2$, we have quadratic or second-order convergence.
In the discussion of bootstrapping variations, I mentioned that the standard algorithm is second-order convergent while subsampling is first-order. Both methods approach the truth as we increase the number of iterations (under certain assumptions), but the standard algorithm approaches faster. Of course, there are places where we might not be able to apply the standard algorithm (we its assumptions are violated), but could be a valid case for subsampling.
We can see this better by comparing error rates in the methods provided in the sanity check above. For now, let’s just look at the full algorithm and subsampling. A plot on logarithmic scales shows the error of the standard algorithm decreasing at roughly twice the rate of that of subsampling.
# Create a range of iterations
i_range <- cross_n(list(x = c(5,10), y = 1:5)) %>%
map_dbl(~ .$x^.$y)
# Test errors across i_range
full_err <- map(i_range, bs_sim, x = x, replace = TRUE, full_funs, n, b = n, mu)
subsamp_err <- map(i_range, bs_sim, x = x, replace = FALSE, red_funs[c(1,2)], n, b = b, mu)
converge_results <- list(standard = full_err, subsamp = subsamp_err) %>%
map(bind_rows) %>%
bind_rows(.id = "sampling") %>%
mutate(method = gsub("^.*_", "", method),
bias.sqr = sqrt((true$lwr - lwr)^2 + (true$upr - upr)^2),
iter = rep(i_range, 2, each = 2))
# Plot
converge_results %>%
ggplot(aes(iter, bias.sqr)) +
geom_line(aes(color = sampling)) +
facet_grid(. ~ method) +
scale_x_log10() +
scale_y_log10() +
ggtitle('Convergence Rates for Standard Bootstrap and Subsampling')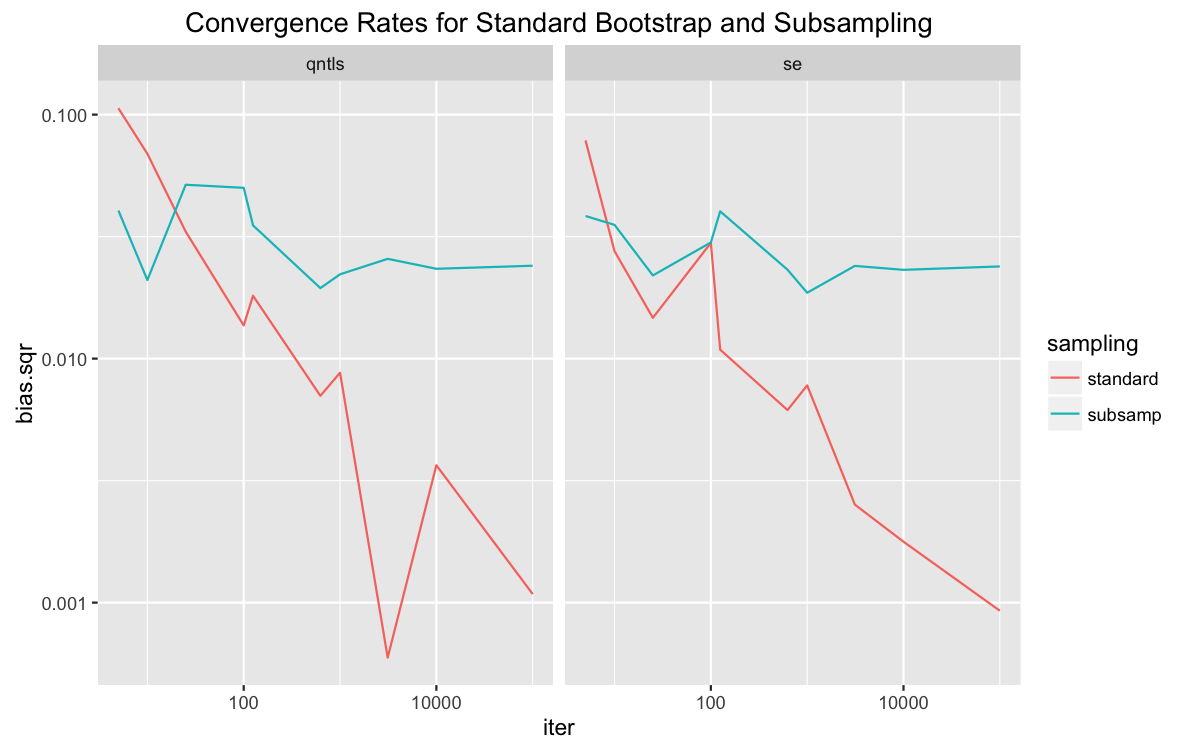
The results aren’t very smooth, which is evidence that the normal approximation in the plot on the right doesn’t work well with the number of samples is small. Nonetheless, the error does decrease as the number of iterations increases. The rate for the full method is much greater, which is what the theory explains.
Running the experiment
Since our functions are stored in a list, calling the experiment is really simple. We’ll just use the default values for all of the test parameters. Remember, we are always sampling less than the full data. The key difference is how much and how we’re correcting for this bias when calculating confidence intervals.
# Simulate replacement bias -----------------------------------------------
test <- bootstrap_experiment(red_funs)We can visualize the results with a boxplot. The columns facet each type of bias measured, while the rows facet if replacement was used in sampling.
# Box plot
test %>%
gather(bias_type, value, bias.lwr, bias.upr, bias.sqr) %>%
ggplot(aes(method, value)) +
geom_boxplot(outlier.shape = 1) +
facet_grid(replace ~ bias_type, scales = 'free') +
ggtitle('Box plot of biases for each method')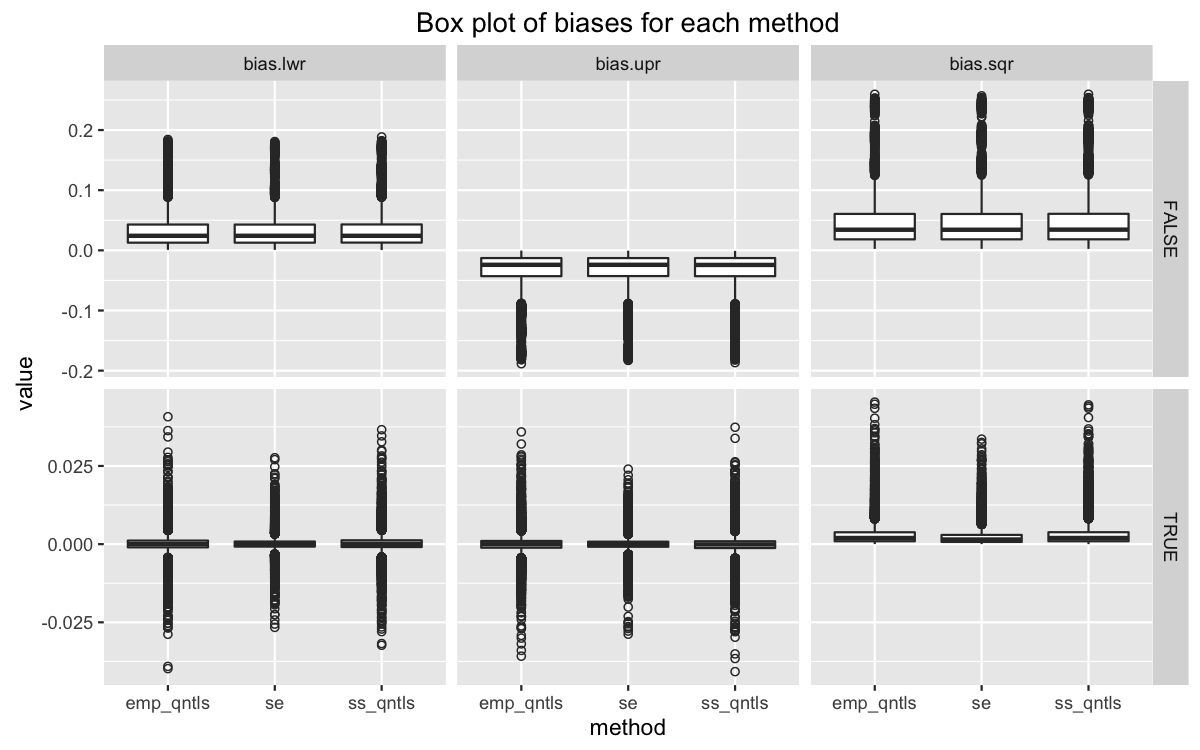
When replacement is used, the expected values upper and lower bias is very close to zero. The 2-norm is slightly greater than zero. Again, this convergence rate is an order of magnitude higher than that for instances where replacement is used. We see the same in a simple summary table.
While both methods might be converging, we have some limited evidence to say the m out of n bootstrapping is doing a better job of eliminating errors. A model will give us a better look at convergence rates for each method.
An analysis of variance
We’ll take a deeper look at convergence rates by implementing an analysis of variance. To do this, we start by comparing all first-order effects and interactions, and then trim accordingly. This method says we should not include the mean, as it doesn’t affect bias in a significant manner. A couple of interactions with the method of calculating the confidence intervals can also be dropped.
To spare you the some of the tedium, I’ll only present the ANOVA table for the remaining elements. Here it is.
# Anova, convert test levels to factors
reshape <- test %>%
select(bias.sqr, method, n, r, sigma, bsi, replace) %>%
mutate(method = factor(method, levels = c("ss_qntls", "emp_qntls", "se")))
model <- aov(bias.sqr ~ (.) * (.) - method:r - r:bsi, reshape)
summary(model)## Df Sum Sq Mean Sq F value Pr(>F)
## method 2 0.00 0.00 1.372e+01 1.10e-06 ***
## n 1 9.77 9.77 6.833e+04 < 2e-16 ***
## r 1 8.77 8.77 6.136e+04 < 2e-16 ***
## sigma 1 13.57 13.57 9.490e+04 < 2e-16 ***
## bsi 1 0.04 0.04 2.787e+02 < 2e-16 ***
## replace 1 44.17 44.17 3.090e+05 < 2e-16 ***
## method:n 2 0.00 0.00 2.514e+00 0.08099 .
## method:sigma 2 0.00 0.00 3.228e+00 0.03964 *
## method:bsi 2 0.00 0.00 2.935e+00 0.05313 .
## method:replace 2 0.00 0.00 6.544e+00 0.00144 **
## n:r 1 1.51 1.51 1.058e+04 < 2e-16 ***
## n:sigma 1 2.35 2.35 1.642e+04 < 2e-16 ***
## n:bsi 1 0.01 0.01 4.822e+01 3.84e-12 ***
## n:replace 1 7.61 7.61 5.325e+04 < 2e-16 ***
## r:sigma 1 2.09 2.09 1.464e+04 < 2e-16 ***
## r:replace 1 8.86 8.86 6.201e+04 < 2e-16 ***
## sigma:bsi 1 0.01 0.01 7.467e+01 < 2e-16 ***
## sigma:replace 1 10.57 10.57 7.392e+04 < 2e-16 ***
## bsi:replace 1 0.03 0.03 1.774e+02 < 2e-16 ***
## Residuals 93725 13.40 0.00
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1A good step from here is to produce effects tables. It’s not practical to print all of them here, since so many factors are significant, but here is one of the more interesting results.
tables <- model.tables(model, "effects")
tables$tables$`r:replace`## replace
## r FALSE TRUE
## 0.25 -0.013752010 0.013752010
## 0.375 -0.006876005 0.006876005
## 0.5 0.000000000 0.000000000
## 0.625 0.006876005 -0.006876005
## 0.75 0.013752010 -0.013752010When we don’t use replacement, which is the proper subsampling method, smaller subsamples relative to the original data reduce bias. We can also approach this by plotting the main effects. Since we’re interested in how these effects vary across sampling schemes, we better make sure that we split the predictors accordingly.
model %>% model.matrix %>%
as.data.frame %>%
select(4:8) %>%
mutate(fitted = fitted(model)) %>%
gather(variable, predictor, -fitted, -replaceTRUE) %>%
rename(replace = replaceTRUE) %>%
mutate(replace = as.factor(ifelse(replace == 1, "true", "false"))) %>%
group_by(predictor, replace) %>%
mutate(av_fitted = mean(fitted)) %>%
ggplot(aes(predictor)) +
facet_grid(replace ~ variable, scales = 'free') +
geom_point(aes(y = fitted, color = variable)) +
geom_line(aes(y = av_fitted), color = "black") +
theme(legend.position = "none") +
ggtitle('Fitted values against predictors')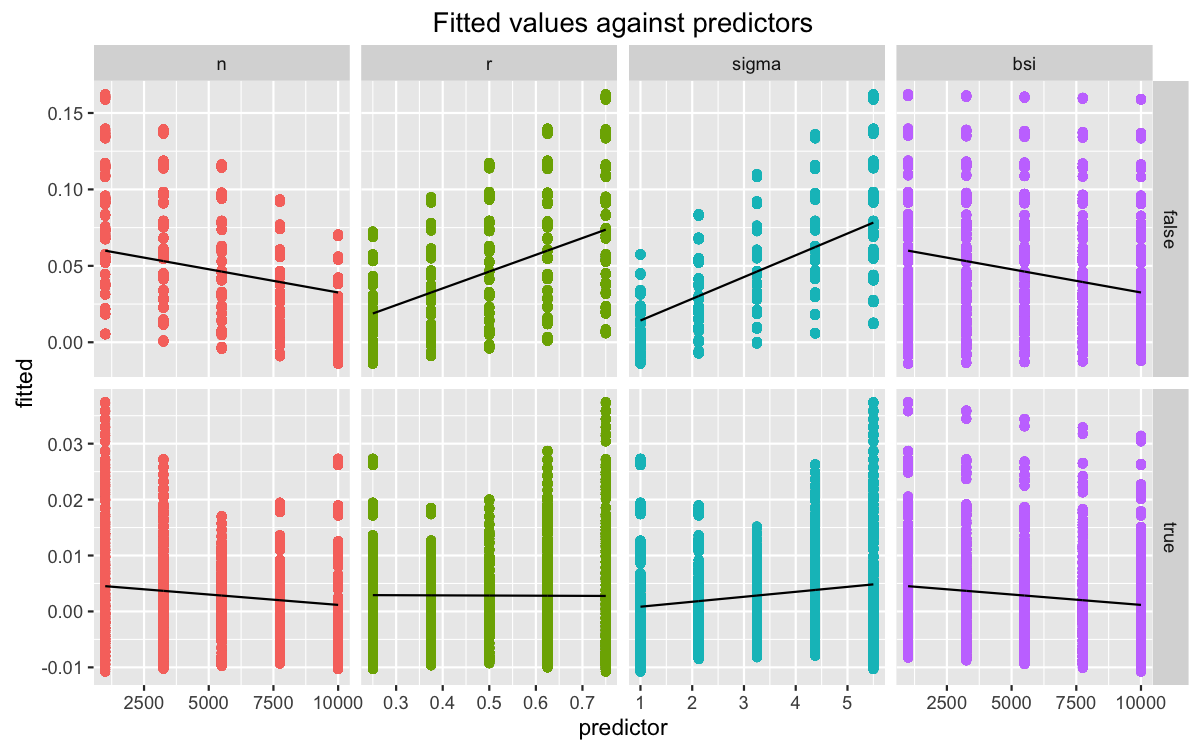
Conclusions
We started off by asking the question: how should you bootstrap? Well, it turns out that there might be many different sampling schemes that fall in the bootstrapping family, and the standard algorithm is only one member of this family. Chernick’s book is a good resource for the many varieties, and separating which methods best suit which problems is a challenge unto itself. Looking at just two of those methods, subsampling and the m out of n bootstrap, we see that a variety of parameters influence the size of the bias in confidence intervals.
The best combination of parameters is quite tricky. As Bickel and Sakov note, choosing the size of the subsample is in fact one of the hardest steps in using these methods. We don’t resolve these problems here to any degree of certainty, but there are some general practical implications. Subsampling seems to prefer smaller subsamples relative to the size of the original data, and both methods improve as the size of the original data and the number of iterations increase.
Of course, be sure to use common sense in reducing the size of the subsample. If your sample is too small, certain methods for calculating confidence intervals become suspect at best or entirely invalid at worst. The size of your subsample needs to be considered against the size of your original data as well. Unfortunately, this means that simple heuristics like 100 observations or 10 percent don’t really apply. You need to experiment and iterate to get better results.
I hope you enjoyed this limited tour of these methods, and please don’t be afraid to provide feedback!
-
They should be, but that’s a rant for another day. ↩

